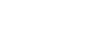Embryonic Development
LifeMap Discovery® as a Novel Portal for Mammalian Developmental Biology
LifeMap Discovery provides easy access to a wealth of information on mammalian development and integrates it with stem cell biology and regenerative medicine information.
- A detailed description of the developmental ontology of organ/tissues, anatomical compartments and cells
- Manually curated gene expression at all developmental stages, as well as data extracted from high throughput experiments and large scale in situ databases
- Detailed information on signals that regulate cells during development
- A roadmap for in vitro cell differentiation by matching cells of interest to in vivo entities
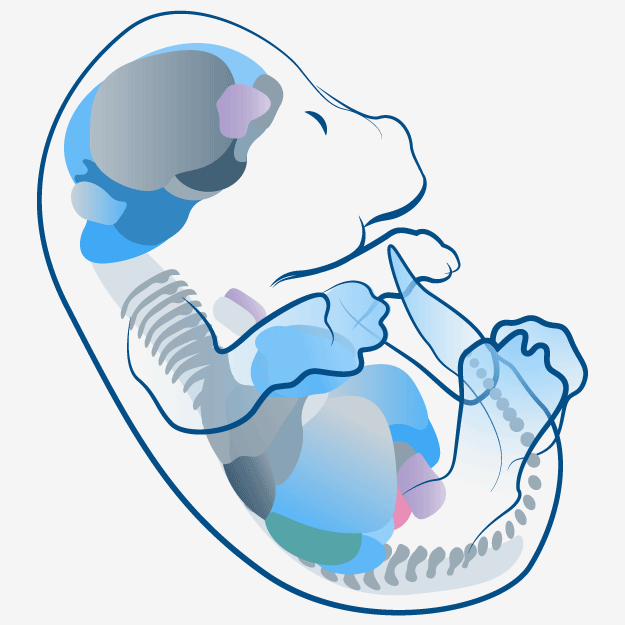
Embryonic Development Modeling in LifeMap Discovery
The Tri-leveled Structure
LifeMap Discovery captures the cellular differentiation that occurs during mammalian embryonic development, as well as the concerted differentiation events leading to formation of anatomical compartments within complex organs and tissues. The database covers multiple developmental paths and provides a snapshot at cellular and anatomical resolutions. The complexity of the developing embryo is approached by classification (tags) used to organize the data types (entities) in relation to each other, as defined below:
"System": A collection of organs, which share similar functions. The system tag provides a physiological context to organs and tissues. For example, the reproductive system, which includes the ovary and testis organs.
"Organ": A collection of tissues and specialized cells joined in a structural unit to serve common sets of functions. The organ tag provides an anatomical and functional context to its populating anatomical compartments and cells.
"Tissue": A collection of cells that, together, perform specific functions, although they can be found in multiple anatomical regions. The tissue tag provides a functional context to its related anatomical compartments and cells.
"Developmental path": the course of cellular development, from the cell's first ancestor(s), termed a “primary progenitor”, to the fully differentiated cell. The developmental path tag provides a developmental context for the cell, linking it to its potential developmental fates and enabling longitudinal analysis along the developmental ontology tree. A cell's specific temporospatial localization within an organ or tissue does not always dictate its functional fate, as demonstrated by hematopoietic stem cells located in the bone marrow, which are anatomically related to bone, but belong to the “blood” developmental path. Similarly, dopaminergic neurons constitute a developmental path of specific neural cells in the brain.
"Cell class": cells that perform specific functions and express common selective marker genes, although they may originate from different ancestors, are located in multiple anatomical regions, and can belong to different developmental paths. For Example: mature choroid plexus cells that are located in the brain belong to the cell class epithelium.
The Organ/Tissue Level
The Organ/Tissue level provides a low-resolution description of mammalian development, as well as related stem cell differentiation and regenerative medicine information. Entity types at this level include: System, Organ, Tissue and Developmental path (see definitions of each tag type in the previous chapter). They are organized in the Embryonic Development Ontology Tree, beginning at the zygote, proceeding via the three germ layers and their extraembryonic components, and extending to fully developed organs and tissues that comprise the adult body. Each entity has a corresponding “card”, which provides a general description of its function, structure and developmental path, along with gene expression and signaling data. The card contains an illustration of the entity, as well as links to related anatomical compartments, cells, differentiation protocols, related stem cells, diseases and cell therapy applications within LifeMap Discovery.
The Anatomical Compartment Level
The anatomical compartment level outlines the development of specific temporospatial regions within an organ/tissue. At this level, each entity is linked to its related organ/tissue and can be linked to one or more parent compartments. The anatomical compartment developmental tree begins with a parent compartment, which can give rise to one or more compartments.

A descendant compartment is presented in the viewer as “developed-from” its ancestor, indicated by a white line connecting two anatomical compartments. In addition, a descendant which is merely a part of another anatomical compartments is presented as a “part of” its ancestor, indicated in the viewer by an orange line connecting two anatomical compartments.
Each anatomical compartment has a corresponding card that provides a brief description, accompanied by images describing its structure, anatomy and development, as well as links to its populating cells. In addition, the card provides lists of genes specifically expressed in the anatomical compartment or in its populating cells, as annotated from large scale in situ hybridization databases, and relevant high throughput experiments. The card also provides links to related stem, progenitor and primary cells, as well as details of associated diseases and cell therapy applications related to the anatomical compartment or to its populating cells.
The Cell Level
The cell level details differentiation paths taken by cells directed toward specific lineages, are studied
in mouse, human or chicken. Each cell is mapped as a descendant of one or multiple ancestor cells, and
is linked to an anatomical compartment, based on its final location. In turn, each anatomical compartment
is linked to an organ/tissue, generating a unique nomenclature (Embryonic Index - EIndex) which distinctly
identifies the cell within the anatomical context of the developed embryo.
Each cell has a corresponding card that describes the developmental stage, and provides a list of expressed
genes which was gathered by manual curation and/or extracted from related microarray and large scale
in situ hybridization experiments, as well as signaling and signal transduction cascades. Additionally,
each cell card contains links to related stem cells and regenerative medicine sections of the database,
as well as links to diseases that affect the given cell or which can potentially be treated by related
in vitro cells.

The information provided in the cell cards aims to assist stem cell researchers to match and identify stem cell derivatives, obtained from differentiation protocols, with embryonic in vivo cells by means of gene expression profiles and selective gene markers. The information provided may also assist researchers in enhancing differentiation protocols, based on available signaling information.
The Ontology Tree Structure
The interactive ontology viewer in LifeMap Discovery is designed to enable researchers to learn about the hierarchical order of development, within the spatiotemporal context of each developing tissue/anatomical compartment/cell. Each entity is connected, via a navigable link, to its ancestor(s) ("developed from") and its descendants ("develops to"). All compartments and cells are also linked to their related organ/tissue, where all the information is aggregated.
Developmental Time
LifeMap Discovery provides annotations in the cell cards for the developmental stage of cells in the following fashion:
DPC (days post-coitum) – Embryonic days (e.g., E13.5 for 13.5 dpc). Human gestational weeks are sometimes added as a note in the cell card of a mouse cell, and mouse developmental times are sometimes annotated in the notes field of human cells.
Stage - numbers representing TS (Theiler Stage), when mouse development is being described, CS (Carnegie stage), when human development is being described, HH (Hamburger-Hamilton) when chick development is being described.
When the cells are present postnatal, the exact days are also annotated (e.g., P10). When the exact developmental time is not available, the timing of first appearance of the cells is generally annotated as either prenatal/postnatal. When information for the developmental time is completely absent, the N/A (not available) annotation is presented. Thus, the developmental time annotations enable us to describe complex multi-staged developmental processes without losing context.
Species Selection
LifeMap Discovery aims to present a carefully compiled and comprehensive view of mammalian development, which is a complex and well-orchestrated process. While some developmental stages are conserved among most mammalian species, significant interspecies variations in developmental processes require a species-specific description of development.
The organ/tissue and anatomical compartment levels do not have a species annotation since information from multiple species can be aggregated to them. However, all the cells in the database have a species annotation in the form of an icon depicting the species in which the provided information was observed.

|
The developmental ontology tree presents a low-resolution overview of the development of the organs/tissues. The "early development" stages, starting from the zygote, through the three germ layers (ectoderm, mesoderm and endoderm), describe human development in the database. Later stages of embryonic development, from the formation of the three germ layers to their derived organs and tissues, are described mostly for the mouse embryo in the database. |
Due to the wealth of information available on early human development, and the clinical relevance of human embryonic stem cells (obtained from human blastocysts) the "early development" stages presented in the database describe human stages, defined as the stages from the zygote to the three germ layers (endoderm, mesoderm and ectoderm). Due to ethical and practical limitations, detailed developmental information for later stages of the human embryo development is scarce. Therefore, the later stages of embryonic development, from the formation of the three germ layers to birth, are described mostly for the mouse embryo, which has been extensively studied by developmental biologists and stem cell researchers worldwide. In some rare cases, information from chick species is provided.
Species type is also annotated for gene expression, and appears as an icon near the gene name. The logic behind presentation of species-diverse gene expression information is:
- A lot of data on gene expression during development belongs to species whose development is not described in our database, thus the information is not lost and is included in the relevant mouse/human cells.
- Often, information from the literature is partial and the ortholog is not known.
Signal Modeling
Details of development-associated signals are outlined in the database, and signal cascades are described in depth. The described signals are generally the more central signals that define the progress of the differentiation process along the developmental path. Careful rules have been defined, as each signal described in the literature must be carefully interpreted with regards to the cell receiving it and the cell affected by it, defining the outcome to the incoming signal.
Incoming, Internal and Outgoing Signals
Signals that are activated by secreted proteins are defined in LifeMap Discovery as "incoming signals" and are detailed in the cell cards relating to cells that are affected by the signal. When the cells that secrete the signal are known, they are mentioned in the signal details as the source cells. In such cases, the signal is presented in the source cell card as an "outgoing signal". Often, the actual secreted molecule that activates the signal is unknown, while its effect, e.g., induction or inhibition of certain genes, such as transcription factors, is known. Such signals are defined in the database as "internal signals" and their name depicts the downstream genes affected by the signal, followed by their expression effect (as 'induced' or 'inhibited').
Gradient Signal
If the secreted signal arrives to the cell as a gradient, and a specific gradient level has been shown to be critical in determining the effect on the cell, the level of the gradient is annotated (low, low-medium, medium, medium-high, high). Effects triggered by one molecule but at different gradient levels are presented as separated signals.
Genes in Downstream Cascade
Each signal affects genes or gene products. When the affected genes are known, the signal cascade is presented, where the effect is defined as: induced, inhibited or involved. A gene that is labeled as "Involved" in a signal means that its product is involved in molecular interactions with other genes products within the cascade, although its own gene expression (or activity) remains unaffected by the signal.
For each signal, the genes known to participate in the downstream cascade, and the relationship between them, can be viewed under "see details" within the signal scheme.
Signal Outcome
Several types of signal outcomes are described:
-
- Differentiation:
- A process in which a less specialized cell becomes more specialized or takes on a more determined fate.
-
- Epithelial-to-Mesenchymal Transition (EMT):
- A process in which epithelial cells lose their cell polarity and cell-cell contacts, resulting in increased migratory and invasive potential and generation of mesenchymal cells.
-
- Mesenchymal-to-Epithelial Transition (MET):
- A process in which motile mesenchymal cells gain cell polarity and cell-cell adhesion properties and eventually become stationary epithelial cells.
-
- Proliferation:
- A process in which cells multiply and increase in number.
-
- Migration:
- A process in which cells move from one region to another.
-
- Apoptosis:
- A cellular death process in which cells undergo a programmed sequence of events, marked by the fragmentation of nuclear DNA, without releasing harmful substances into the surrounding area.
-
- Inhibition of Differentiation:
- A process in which the signal prevents the less specialized cell to differentiate to a specific fate.
-
- Cell Cycle Arrest:
- A process by which the mitotic cycle is halted.
-
- Survival:
- A process enabling the continued existence of a cell. This type of signal effect is common to neurons.
-
- Maintenance:
- A process in which cells retain their cell identity.
If the signal affects the cell by inducing differentiation or another processes which result in a change in cell identity (EMT, MET, Migration, or Inhibition of Differentiation), a descendent cell ('derived cell') will be linked to the signal.
Matching to Stem Cells
Stem cells serve as a platform for extensive research largely aimed toward clinical applications, such as cell replacement therapeutics, trophic support and in vitro disease models. Each stem cell procedure normally follows specific protocols which describe the growth conditions and cell markers for these cells. Often, the stem cells used as source cells for the protocols are difficult to characterize, and as the cells differentiate, cell identity is difficult to establish and is frequently based on few cell surface markers, fragilely linking them to known lineages (and occasionally, uncharacterized side populations of cells remain after the differentiation process, contaminating the otherwise homogenous culture).
The ability to match differentiating stem cells to their counterparts in live, developing organism, offers the researcher powerful tools for characterizing and understanding the differentiation process.
Understanding the molecular signals and gene expression patterns that regulate cellular differentiation and maturation, can further enhance design of differentiation protocols that better mimic in vivo processes.
The underlying assumption motivating the matching effort is that stem or progenitor cells undergoing in vitro differentiation show similar gene expression patterns to their equivalent in vivo counterparts. In line with this reasoning, knowledge of the exact or even approximate in vivo correlate of a cell, i.e. its position within the developmental ancestry tree and its associated anatomical location or tissue type classification, can be extremely useful.
LifeMap Discovery exploits the wealth of data accumulated on gene expression in specific cells during development, to match in vitro cells to one or more in vivo cells or anatomical compartments of the embryo or mature mammal. The matching is performed either manually or computationally, and is primarily based on gene expression profiles, but can also be based on function and morphology observations described in the literature.
Selected Topics in Mammalian Developmental Biology
All mammalian embryos begin their development through the same basic process. The zygotes and early embryos of mammals are virtually indistinguishable from each other until the embryo begins to assume its species-specific characteristics as it develops specialized organs, tissues and cells
The Zygote
The zygote is the first stage of the early embryo, and is formed upon fusion of the haploid (n) male and female pronucleus, wherein the chromosome number in the fertilized ovum becomes diploid (2n). One or two polar bodies are often visible in the perivitelline space, the area between the embryo and the zona pellucida (a glycoprotein membrane that surrounds the plasma membrane of an oocyte).
Fertilization, which normally takes place in the ampulla of the uterine tube, requires contact of spermatozoa with the zona pellucida of the secondary oocyte, penetration of one or more spermatozoa through the zona pellucida and formation of the male and female pronuclei. Completion of the second meiotic division (the first meiotic division occurs prior to fertilization and the first polar body is a result of this division) marks the commencement of the first mitotic division, or cleavage, of the zygote.
Cleavage and Blastocyst Formation
While the zygote is transported through the Fallopian tubes to the uterus, its cells undergo subsequent mitotic divisions, called cleavages, wherein the daughter cells are smaller than the parent, thus, the embryo increases its cell numbers without increasing its total mass or size. The 2, 4 and 8-cell embryo (also known as the early morula), is formed and at this stage the embryonic cells undergo compaction, forming the morula. The mammalian embryo obtains the majority of its nourishment through the placenta and umbilical cord, and therefore, the mammalian cleavage is known as holoblastic cleavage, as opposed to meroblastic cleavage in embryos that receive the majority of their nourishment through an egg yolk. The cells in cleavage-stage embryos are known as blastomeres. As they continue to divide, the cells segregate to the inner and outer cells, forming a cavity (the blastocoele cavity), thereby defining the blastocyst-stage embryo. The inner group of cells (called the inner cell mass, ICM) will become the embryo, while the outer group of cells, called the trophoblast, will become specialized extraembryonic membranes, adapted for intrauterine mammalian development ( chorion, amnion, placenta, yolk sac, umbilical cord). The blastocyst stage is therefore the first time that two distinctive tissues are present, as a result of a segregation that is often described as the first lineage-choice of the embryonic cells.
Trophoblast and the Inner Cell Mass
The blastocyst stage includes dramatic events such as, hatching of the blastocyst from the zona pellucida, and implantation of the embryo in the uterine wall. The zona pellucida prevents adherence of the blastocyst to the oviduct walls (a condition which in humans is called an ectopic ortubal pregnancy). When the embryo reaches the uterus, zona hatching enables implantation, a complex series of events that involves the formation of the enveloping syncytiotrophoblast layer and cytotrophoblast layers of the placenta. Both layers originate in the embryonic trophoblast, and invade the maternal decidua (the maternalpart of the placenta), creating an extensive interface with the maternal tissues.
The epiblast is formed as the ICM segregates into a bilaminar embryonic disc (bilaminar blastoderm), which consists of two epithelial layers: the external (dorsal) epiblast and the internal (ventral) hypoblast. The hypoblast cells delaminate from the inner cell mass to line the blastocoel cavity, where they give rise to the extraembryonic endoderm, which forms the yolk sac.
Germ Layer Formation
Gastrulation is the process in which the ICM is converted into the three germ layers: ectoderm, mesoderm and endoderm. Cells begin to migrate into the center of the blastula and to form primitive germ layers, transforming the blastula into a gastrula. Formation of the primitive streak marks the first event of gastrulation. The primitive streak originates from the anterior epiblast, and appears as an elongating groove (primitive groove) on the dorsal midsagittal surface of the epiblast, along the anterior-posterior axis of the embryo. The rostro-caudal and medial-lateral axes of the embryo are defined by the primitive streak. The rounded primitive node, or Hensen's node, is situated at the cranial tip of the primitive streak, and contains a depression called the primitive pit, which is continuous with the primitive groove. Cells from the epiblast migrate into the interior of the embryo, via the primitive streak, in a process termed ingression, which involves a cellular epithelial-to-mesenchymal transition (EMT). The initial wave of migrating cells streams through the primitive streak, displacing the hypoblast cells to become the definitive endoderm, which ultimately produces the future gut derivatives and gut linings.
The second wave of migrating cells populates a layer between the epiblast and the definitive endoderm, thereby forming the mesoderm layer. The intraembryonic mesoderm cells later give rise to five subpopulations of cells: paraxial mesoderm, intermediate mesoderm, lateral plate mesoderm, cardiogenic mesoderm and a population that forms a midline tube called the notochordal process. The notochordal process originates in the primitive node and is the precursor of the flat-shaped notochordal plate, which after detaching from the endoderm, fuses its free rims together to form a rod that is known as the notochord. Once the mesoderm has formed, the remaining epiblast cells cease to ingress and form the ectoderm. The ectoderm gives rise to two distinct lineages, namely, the surface ectoderm and the neural crest.
Further Development
Cellular specialization continues throughout the gestational period, leading to formation of over 200 tissues and cell types. Somites begin to block off and form the basis for the spinal column, skeletal muscles, vertebrae and dermis in all mammalian embryos. Then, organogenesis begins, involving specialization of organ systems, including the nervous, circulatory, reproductive, sensory and digestive systems. Further growth and maturation of the organs and tissues continues throughout the remainder of the gestational
Central Signaling Pathways
A number of key signaling pathways are responsible for central developmental stages and determine the architecture, patterning and specification of the embryo. Such key Signals are usually highly conserved among species, and mutations or disruption in their cascade result in severe malformations or arrested development. If asked, most embryologists would probably point out the following principal signals in embryonic development: Wnt, Notch, BMPs, FGFs and SHH.

|
Prominent Markers and Signals during Oligodendrocyte development. Signals and gene expression markers characteristic of developing oligodendrocytes derived from the spinal cord and the telencephalon. Signals are indicated alongside the icons representing their effect (see legend at the bottom of the image) and the gene expression markers are indicated in the center of the image, just below cell names. (A) Neuroepithelial progenitor cells within the spinal cord and the telencephalon differentiate into (B) oligodendrocyte precursor cells, which are proliferative and migratory cells that populate the spinal cord and forebrain white matter and later develop into (C) pre-myelinating oligodendrocytes, which mature to become (D) myelinating oligodendrocyte cells that synthesize myelin. |
The Wnt Pathway
Wnt proteins are one of the key families of developmentally important signaling molecules, with mutations in Wnt genes displaying remarkable phenotypes. Among the functions provided by Wnt proteins are intriguing processes such as generation of cell polarity and specification of cell fates. Many different pieces of knowledge from a range of species have converged on three common Wnt pathways, which are usually classified as the canonical Wnt pathway (also known as the Wnt/β-catenin pathway), which controls and modulates gene transcription, the noncanonical planar cell polarity pathway, responsible for modulating the cytoskeleton, and the non-canonical Wnt/calcium pathway, which modulates intracellular calcium levels. While these pathways differ in many ways, all three share a common ligand-associated activation of a frizzled family receptor, which, in turn, activates the intracellular disheveled protein. Wnt pathway signaling plays a key role in numerous developmental steps, including body axis formation, induction of organ formation and cell differentiation, regulation of cell proliferation and migration.
WNT signaling outcomes can be contradictory, as manifested during embryonic skeletal muscle formation. While canonical WNTs (WNT1 and WNT3a) are important in paraxial mesoderm (i.e., somite) induction and differentiation towards muscle cells in the trunk region (axial muscles), the same signals were shown to be inhibitory for head myogenesis.
The SHH Pathway
Sonic Hedgehog signaling is another fundamental signal in embryonic development. Shh pathway signaling involves the ligand-mediated activation of two transmembrane proteins - Patched receptor and Smoothened (Smo) signal transducer. The signal affects the intracellular transcription factor Gli (homologues 1-3), thereby inducing transcription of multiple genes.
This pathway is conserved among species and, when aberrant, causes severe malformations and growth arrest. Sonic Hedghog is one of three mammalian orthologues of the drosophila Hedghog (the other two are Indian Hedghog and Desert Hedghog) and acts in the pathway is involved in regulation of cell fate and embryonic patterning during early vertebrate development and in the formation of multiple organs, such as the heart, lungs, eye, brain, bone, pancreas and spinal cord, among others. The Shh pathway remains active in the adult organism, where it plays a role in maintaining populations of multipotent stem cells.
The Notch Pathway
The Notch signaling pathway, plays a significant role in regulating cell fate decisions throughout mammalian development, affecting neuronal, cardiac, immune, and endocrine development. Notch signaling remains active in the adult, modulating maintenance of adult tissue homeostasis. As befitting a key pathway, the Notch signaling pathway is evolutionarily conserved. The pathway is activated in a juxtacrine system, wherein both the ligand-presenting cell and the receiving cell are affected. Four Notch homologues have been identified in the human genome and aberrant Notch function has been associated with a number of human diseases.
In mammalian Notch pathways, members of the Delta-like Jagged (JAG1, JAG2) and the DLL (DLL1, DLL3, DLL4) families serve as ligands for the Notch receptors. The ligand-receptor complex formation induces a three-series cleavage event, which facilitates downstream effects on transcription via the activation of the canonical Notch target genes: Myc, p21, and the HES-family members
Notch signaling plays a key role in hair follicle cells differentiation, by defining the outer root sheath (ORS) cells. Mutations of Notch family members cause a switch from ORS to epidermal differentiation, resulting in replacement of hair follicle by an epidermal cyst. In addition, during epidermal stratification, Notch signaling plays a key role in inducing differentiation of suprabasal layers. Differentiation of spinous keratinocytes into granular keratinocytes is Notch-dependent and is defined in two-step signaling process: first, Notch1 inhibits differentiation, allowing emergence of a sufficient number of cells by proliferation. When the appropriate number of cells is accumulated, Notch induces cell cycle exit and progress into the differentiation stage.
The BMP Pathway
Bone morphogenetic proteins (BMPs) are members of the TGF-beta superfamily; approximately 20 BMPs have been identified to date. While BMPs may induce ectopic bone or cartilage formation, they also play important roles in many other developmental processes, including cell proliferation, apoptosis, differentiation, and morphogenesis.
BMPs induce the formation of both cartilage and bone. During bone formation, BMPs facilitate the development of functional bone marrow. In addition, BMPs play a role in a number of non-osteogenic developmental processes. BMP-2 induces the neuronal phenotype of neural crest cells, while BMP-4 directs somite development by inhibiting myogenesis. BMP-4 and BMP-7 specifically induce a sympathetic adrenergic phenotype. In the limb bud, and as part of the FGF-4 and SHH interaction, BMP-2 inhibits limb bud expansion and induces the formation of chondrocyte and osteoblast precursor cells. BMPs are also involved in dorsal/ventral patterning. BMP-4 specifies the development of ventral structures (skin from ectoderm and connective tissue/ blood from mesoderm). Competitive inhibition of BMP4 is responsible for the development of dorsal structures (nervous system and muscle). Recent reports indicate that a heterodimer composed of BMP-4 and BMP-7 is a potent inducer of mesoderm.
Types I and II TGF-beta receptors play significant roles in BMP binding and signaling. Upon ligand binding, heterotetramerization occurs, as the type II receptor activates the type I receptor kinase domain, thereby initiating phosphorylation of cytoplasmic Smad proteins and downstream signal transduction.
The FGF Pathway
The FGF signaling pathways are thought to play substantial roles in development, angiogenesis, hematopoiesis, and tumorigenesis. In addition to their mitogenic capacity, FGFs also modulate cell survival, migration and differentiation in culture. During embryogenesis, FGF signaling plays an important role in the induction/maintenance of mesoderm and neural ectoderm, control of morphogenetic movements, anteroposterior (AP) patterning, head mesenchyme proliferation, somitogenesis, myogenesis and development of various organs, such as lungs, liver and heart. FGF pathway malfunction is implicated in several congenital diseases (such as dwarfism) and some types of cancer.
To date, the FGF superfamily consists of 23 members, all of which contain a conserved 120-amino-acid (aa)-long core region which is comprised of six identical, interspersed amino acids. The signaling pathway is executed via one of the four tyrosine kinase FGF receptors (FGFRs). In vertebrates, the extracellular domains of the receptors undergo alternative splicing to generate a vast variety of receptors, with variable affinities for their respective FGF ligands. FGF ligands bind the extracellular domain of the FGFR, in combination with heparan sulphate, to form a complex resulting in the transphosphorylation of specific intracellular tyrosine residues, thereby triggering the downstream activation of cytoplasmic signal transduction pathways. Well known signals activated as a result of FGFR activation are the Ras/ERK pathway (affects differentiation), the Akt pathway (affects cell survival) and the protein kinase C (PKC) pathway (involved in cell morphology and migration).