Stem Cell Differentiation
LifeMap Discovery® as a Novel Stem Cell Database
LifeMap Discovery provides information on cultured stem, progenitor and primary cells, along with related differentiation protocols. The integrated knowledge in stem cell biology and embryonic development provided by the database can be harnessed by stem cell researchers to better characterize experimentally obtained cell derivatives and to develop or improve stem cell differentiation protocols.
- Provides valuable information regarding stem cell types
- Contains a large collection of stem cell differentiation protocols
- Enables accurate characterization of cultured stem and progenitor cells during differentiation processes
- Promotes an in-depth understanding of how stem cells differentiate, and of the key signals governing the process
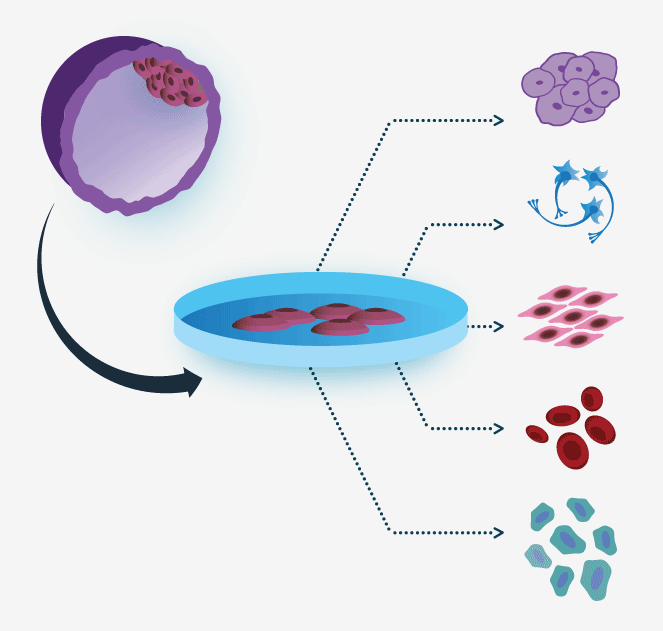
Stem, Progenitor and Primary Cells in LifeMap Discovery
Stem, progenitor and primary cells subdivision provides information on various cells cultured in vitro. Each 'in vitro cell' card provides information regarding essential growth conditions, characteristic gene expression, related cell-therapy applications and relevant references. In vitro cells are matched to one or more in vivo cell(s) or anatomical compartment(s) of the embryo or adult either manually based on function and morphology described in the literature, or based on gene expression profiles by means of a unique biostatistics algorithm developed at LifeMap. This valuable information can assist researchers in choosing cell types most appropriate for specific differentiation protocols and cell therapy applications. In parallel, the presented gene expression profiles and high throughput data can serve as a valuable baseline for classifying unidentified cells obtained under experimental conditions.
Background information
Stem Cell Biology
Stem cells feature two unique characteristics distinguishing them from other cells: their remarkable ability to differentiate into various cell types during early development and in the adult body, and to self-renew, rendering them a robust reservoir of cells for tissue repair. Given these unique properties, stem cells hold great promise for multiple regenerative medicine applications.
During embryonic development, stem and progenitor cells become progressively committed to specific tissue fates. However, while most stem cells differentiate, some stem/progenitor cells are maintained throughout the adult mammalian life. Under exposure to specific triggering signals, these cells will differentiate and replace those which have been lost due to injury or disease or throughout the normal life cycle.

|
Stem cells are characterized by their ability to self-renew and differentiate into specific cell lineages. Differentiation is induced during embryogenesis and in the adult, in response to specific signaling molecules and growth factors. |
Stem cell research aims to develop effective treatments for incurable diseases, complex injuries and chronic wounds. Such applications rely on stem cells to either support undamaged organ components or to replace damaged cells. For example, stem cell-derived pancreatic beta cells can be used to replace damaged insulin-producing beta cells in patients suffering from Type 1 diabetes mellitus. To learn more about stem cell-related cell therapies, see the Regenerative Medicine section in the LifeMap Discovery database.
LifeMap Discovery provides information on two main types of cultured cells:
- Source cells – can be used to initiate differentiation protocols or cell therapies. This category includes embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), embryonic progenitor cells, fetal and adult stem cells and primary cells.
- Protocol-derived cells (PDCs) – cells derived at various stages of multi-step differentiation protocols.
Embryonic Stem Cells (ESCs)
ESCs are derived from pre-implantation-stage embryos donated during in vitro fertilization procedures.
The early embryos are grown to the blastocyst stage, whereupon the inner cell mass is gently removed, placed in appropriate growth medium and cultured on a feeder layer, enabling the cells to propagate and divide. ESCs are pluripotent and can indefinitely proliferate in culture without differentiating or developing genetic abnormalities. Spontaneous differentiation occurs following withdrawal from conditions supporting the undifferentiated state (e.g., feeder cells), resulting in formation of spherical aggregates known as embryoid bodies (EBs).

|
Embryonic stem cells are derived from blastocyst cells obtained during in vitro fertilization procedure. The sperm and egg cells are fused to form a zygote, which is the initial cell of the embryo. The zygote undergoes multiple divisions and after five days in culture the blastocyst is formed. The blastocyst is comprised of an inner cell mass, which subsequently forms the embryo, and a surrounding outer trophoblast layer, which later forms the outer chorionic sac and the fetal component of the placenta. The inner cell mass is isolated and cultured under specific growth conditions to support pluripotency and self-renewal. The resulting cells are now termed embryonic stem cells – ESCs. ESCs can be further differentiated into cells of any of the three germ layers: endoderm, mesoderm and ectoderm. |
EBs contain cellular derivatives of all three germ layers: ectoderm, mesoderm and endoderm. Following exposure to defined conditions, EB-derived cells can further differentiate, into any cell types of the body. The ability of ESCs to give rise to terminally differentiated cells promises to revolutionize medicine by providing an unlimited cell source for transplantations. In addition, these cells are valuable in pre-clinical research evaluating the effects of candidate drugs.
Induced Pluripotent Stem Cells (iPSCs)
Recent discoveries have allowed for reprogramming of adult somatic cells into embryonic-like cells, termed induced pluripotent stem cells (iPSCs). The reprogramming procedure involves introduction of key transcriptional regulators responsible for maintenance of pluripotency and stemness, delivered via viral vectors, as exogenic proteins, microRNAs or episomal vectors.

|
Induced pluripotent stem cells are derived from somatic cells isolated from various tissues and cultured in vitro. The cells are exposed to a combination of transcription factors, the process called 'reprogramming', to achieve pluripotent state. Once reprogramming is completed and the cells express specific markers of pluripotency, the resulting cells are termed induced pluripotent stem cells – iPSCs. iPSCs, similarly to ESCs, are able to differentiate into cells of any of the three germ layers: endoderm, mesoderm and ectoderm. |
The essential transcription factors – Klf4, Sox2, Oct4, and Myc – were first defined and introduced by Shinya Yamanaka, the pioneer researcher in the field. Later, additional combinations of transcription factors that can promote iPSCs induction were discovered by other groups.
The reprogramming technique enables generation of autologous pluripotent cells, minimizing risk for immune rejection when applied for tissue regeneration purposes (Figure 3). Yet, development of safe reprogramming methods which avoid stable genetic modifications will be required before clinical application of iPSCs and their derivatives.
Recently, iPSCs have been applied towards in vitro disease modeling. Patient fibroblasts have been successfully reprogrammed into iPSCs, enabling researchers to study disease-related mechanisms and to perform effective drug screening. In addition, patient-specific iPSCs can be genetically modified to yield healthy cells aimed to be transplanted back to the patient body as a cell therapy to promote recovery. Further information on patient-derived iPSCs can be found in the "Stem, Progenitor and Primary cells" section unit and also in the related disease cards.
Embryonic Progenitor Cells (PureStem™)
PureStem human embryonic progenitor cells (hEPCs) are clonally-purified embryonic progenitor cells derived in vitro from human embryonic stem cells (hESC),
using a "shotgun" method. In this process, hESCs spontaneously differentiate into mixed cell colonies that are subsequently dissociated into single-cell
suspensions and are clonally proliferated on a defined matrix for varying periods of time and under different growth conditions.
This strategy yields hundreds of distinct types of stable non-tumorigenic clonal hEPCs.
Each clone of PureStem hEPC represents homogenous cell population, limiting the occurrence of contaminating cell types
during differentiation that can appear when using pluripotent stem cells. Unlike adult stem cells, they are highly proliferative and have been proven to
create cells and tissues that cannot be produced using adult stem cells. These characteristics make them ideal candidates for regenerative medicine and stem cell research.
High-throughput microarray analyses revealed a unique set of gene markers, surface antigens and growth
factors for each hEPC, allowing for simple matching to specific cells in developmental lineages.
PureStem cell lines are commercially available at
ESI-BIO.
Innovative Human Stem & Progenitor Cell Reagents at ESI-BIO
Fetal and Adult Stem/Progenitor Cells
Adult or fetal stem cells, also called somatic stem cells, are undifferentiated cells found in adult or fetal tissues, capable of undergoing self-renewal and differentiating into specialized cell types. Adult stem cells are generally found in a quiescent (non-dividing) state until they are activated by specific physiological conditions, which can include disease or tissue injury. Although their differentiation capacity is limited compared to ESCs or iPSCs, the ability to isolate them directly from patients makes them attractive candidates for personalized regenerative medicine approaches.
Primary Cells
Primary cells are isolated from adult tissues, possess limited propagation capacity in vitro and have no identified differentiation potential. These cells are commonly used:
- As feeder support for differentiation of stem cells (see LifeMap Discovery Differentiation Protocols)
- As a source for cell therapies (see LifeMap Discovery Cell Therapies)
- As a baseline for analysis and characterization of novel cells in high throughput and gene expression profiling studies
Tissue/Cell Composite
Artificially engineered tissue constructs comprised of cells cultured on a matrix or polymeric scaffold are described in this section. The constructs are used for cell therapy applications and are designed in attempt to develop three-dimensional products for tissue repair.
Cell Families
In LifeMap Discovery, stem, progenitor and primary cells sharing a common origin, molecular characteristics and mode of derivation are grouped into cell families. Each Cell Family card includes a short description of the family, along with a list of all family members available in the database and relevant cell therapies. In addition, each Cell Family card includes an extensive list of genes expressed in at least one of the family members.
The main stem cell families in the LifeMap database are:
Protocol-Derived Cells (PDCs)
Protocols-derived cells (PDCs) are cells derived at various stages of multi-step differentiation protocols and characterized by gene expression and functional assays, PDCs, protocol description and all available cell characteristics and are presented in protocol cards.
Stem Cell Differentiation Protocols in LifeMap Discovery
The Differentiation Protocols section in LifeMap Discovery provides a collection of detailed procedures used to induce differentiation of ESCs, iPSCs and adult stem or progenitor cells. Scientific papers describing the protocols are selected based on their novelty and precision in characterization of the resulting cells, where the highest priority is given to publications in the most distinguished scientific journals. Differentiation protocols are categorized based on association of the derived cells to a specific organ/tissue (e.g., adipose, blood, bone, brain, eye, heart, pancreas, spinal cord).
Background information
Growth factors and small molecules for differentiation induction
Stem cell differentiation is induced when signaling pathways maintaining pluripotency and self-renewal are down-regulated enabling the cells to respond to differentiation signals. Spontaneous differentiation can be achieved by removal of ESCs from their feeder cell layer and/or culturing them as EBs. However, spontaneous differentiation produces a mixture of immature differentiated cells, and the resulting final yield for each specific cell type is very low. Researchers use growth factors, known to modulate specific signaling pathways, in attempt to mimic embryonic development and enhance differentiation outcome. For example, retinoic acid (RA) signaling implicated in neurogenesis and diversification of motor neurons during embryonic development has been shown to promote neuronal differentiation in vitro when added to stem cell cultures.
Recently developed synthetic small molecules, serving as inhibitors or inducers of specific signaling pathways, have been shown to induce more robust and specific differentiation compared to naturally-occurring growth factors. For example, inhibition of TGFβ signal transduction is possible by blocking the activity of the ligands participating in the signaling cascade. However, considering the high structural variability of the ligands belonging to TGFβ family (TGFs, BMPs, GDFs), multiple blocking molecules (Noggin, Follistatin, Chordin) should be used to achieve broad inhibitory effect. Using small molecule inhibitor SB-431542 proves advantageous since it simultaneously blocks three type 1 TGFβ receptors, ALK5, Alk4 and ALK7, preventing signal transduction via transforming growth factor-beta (TGFβ) signaling cascade regardless the nature of the ligand.
Mapped Protocols
These cards contain an informative and interactive viewer sketching the protocol in a step-by-step fashion. Each step has a concise description including duration, growth conditions, selection procedures and relevant functional assays. Each protocol card defines the source cells used for protocol initiation. In addition, at each stem cell type can be matched to one or more in vivo cells in the database based on gene expression profiles by means of a unique biostatistics algorithm developed at LifeMap , or based on manually curated information relating to cell function and/or morphology described in the literature.
Unmapped Protocols
These cards contain a brief description of differentiation procedures that are linked to fully mapped protocol cards within LifeMap Disovery, describing similar methods.